 全部商品分类
全部商品分类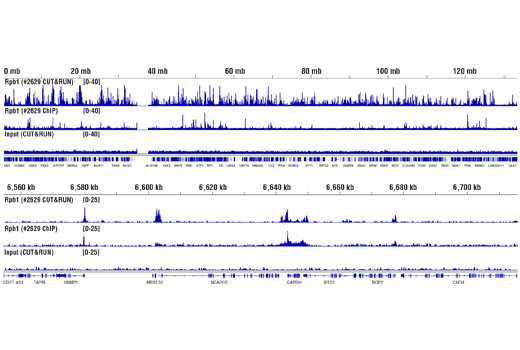



 下载产品说明书
下载产品说明书 下载SDS
下载SDS 用小程序,查商品更便捷
用小程序,查商品更便捷


 收藏
收藏
 对比
对比 咨询
咨询The CUT&RUN Assay Kit is designed to conveniently provide reagents needed to perform up to 8 (P size) or 24 (S size) digestion reactions from cells and is optimized for 100,000 cells per reaction. The kit has been optimized to work for all types of DNA binding proteins, including histones, transcription factors and cofactors. A complete assay can be performed in as little as one day.
The CUT&RUN Assay Kit also provides important controls to ensure a successful CUT&RUN experiment. The kit contains a positive control Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 and a negative control Rabbit (DA1E) mAb IgG XP® Isotype Control (CUT&RUN) #66362, both of which can be used for qPCR or Next Generation sequencing (NG-seq) analysis. PCR primer sets are provided for the human (#7014) and mouse (#7015) RPL30 gene locus to be used in conjunction with the control antibodies. This kit is compatible with both qPCR and NG-seq.


Specificity/Sensitivity



All components in this kit are stable for at least 12 months when stored at the recommended temperature.
参考图片
Figure 12. CUT&RUN and ChIP assays were performed with HeLa cells and Rpb1 CTD (4H8) Mouse mAb #2629, using this CUT&RUN Assay Kit or the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA Libraries were prepared using DNA Library Prep Kit for Illumina® (ChIP-seq, CUT&RUN) #56795. The upper panel compares enrichment of Rpb1 across chromosome 12, while the lower panel compares enrichment at the GAPDH gene, a known target of Rbp1. The input tracks are from the CUT&RUN input sample.
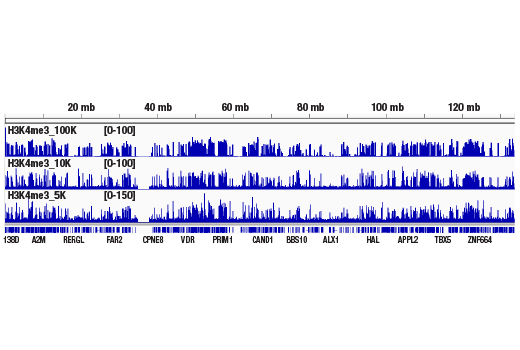
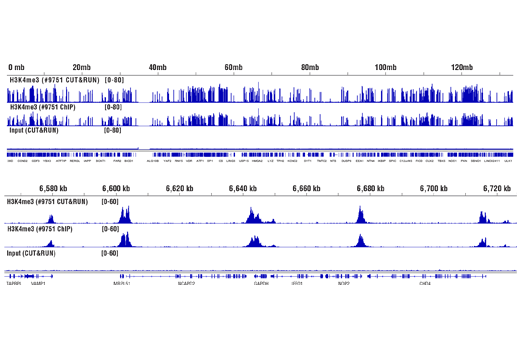
Figure 1. CUT&RUN was performed with 100,000, 10,000, or 5,000 HCT 116 cells (as indicated) and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751, using the CUT&RUN Assay Kit. DNA Libraries were prepared using DNA Library Prep Kit for Illumina® (ChIP-seq, CUT&RUN) #56795. This figure shows binding of H3K4me3 across chromosome 12.
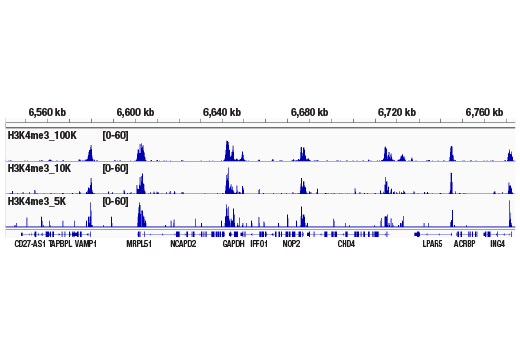
Figure 2. CUT&RUN was performed with 100,000, 10,000, or 5,000 HCT 116 cells (as indicated) and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751, using the CUT&RUN Assay Kit. DNA Libraries were prepared using DNA Library Prep Kit for Illumina® (ChIP-seq, CUT&RUN) #56795. This figure shows enrichment around the GAPDH gene, a known target of H3K4me3.
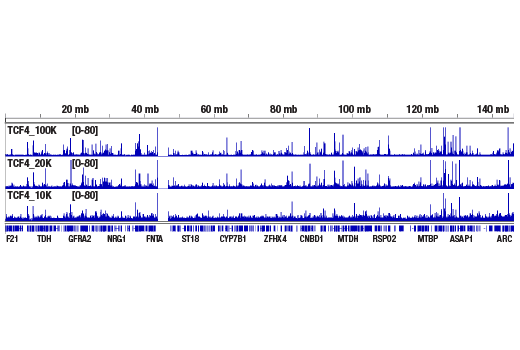
Figure 3. CUT&RUN was performed with 100,000, 20,000, or 10,000 HCT 116 cells (as indicated) and TCF4/TCF7L2 (C48H11) Rabbit mAb #2569, using the CUT&RUN Assay Kit. DNA Libraries were prepared using DNA Library Prep Kit for Illumina® (ChIP-seq, CUT&RUN) #56795. This figure shows binding of TCF4/TCFL2 across chromosome 8.
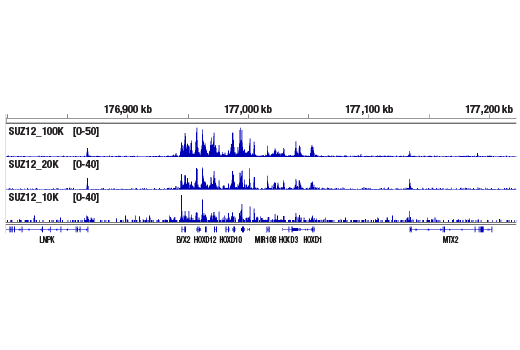
Figure 4. CUT&RUN was performed with 100,000, 20,000, or 10,000 NCCIT cells (as indicated) and SUZ12 (D39F6) XP® Rabbit mAb #3737, using the CUT&RUN Assay Kit. DNA Libraries were prepared using DNA Library Prep Kit for Illumina® (ChIP-seq, CUT&RUN) #56795. The figure shows binding across the HoxD gene cluster, a known target of SUZ12.
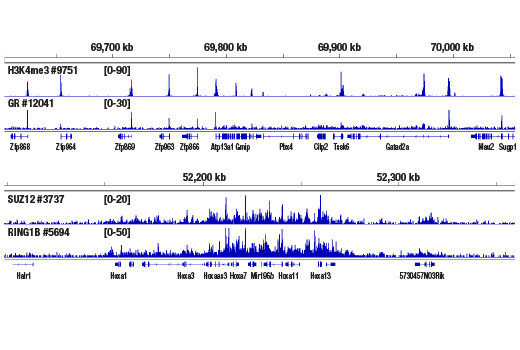
Figure 5. CUT&RUN was performed with 2.5 mg of medium fixed mouse liver tissue (0.1% formaldehyde, 10 min) and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751, Glucocorticoid Receptor (D6H2L) XP® Rabbit mAb #12041, SUZ12 (D39F6) XP® Rabbit mAb #3737, or RING1B (D22F2) XP® Rabbit mAb #5694, using the CUT&RUN Assay Kit. DNA Libraries were prepared using DNA Library Prep Kit for Illumina® (ChIP-seq, CUT&RUN) #56795. The upper panel shows binding of H3K4me3 and Glucocorticoid Receptor around the Zfp866 gene, while the lower panel shows binding of SUZ12 and RING1B across the HoxA gene cluster.
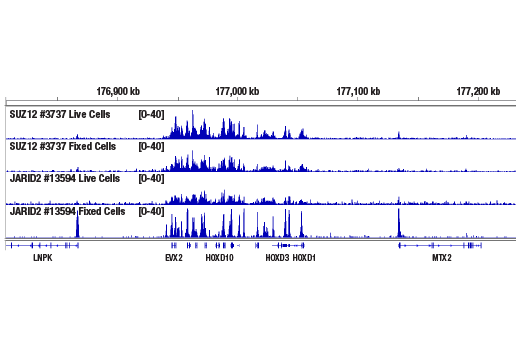
Figure 6. CUT&RUN was performed with 100,000 live or fixed NCCIT cells and SUZ12 (D39F6) XP® Rabbit mAb #3737 or JARID2 (D6M9X) Rabbit mAb #13594, using the CUT&RUN Assay Kit. DNA Libraries were prepared using DNA Library Prep Kit for Illumina® (ChIP-seq, CUT&RUN) #56795. The figure shows binding of SUZ12 and JARID2 across the HOXD gene cluster. Fixation is not required for SUZ12 (a core component of PRC2 complex) but increases the enrichment for JARID2 (an accessory component of PRC2 complex).
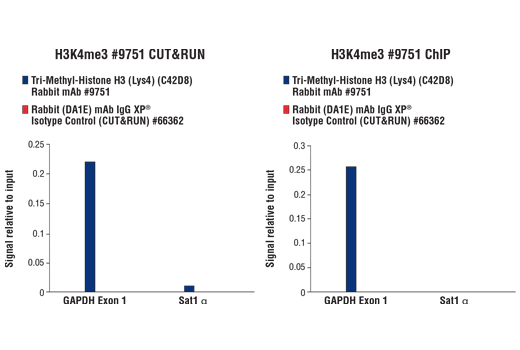
Figure 7. CUT&RUN and ChIP assays were performed with HCT 116 cells and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 or Rabbit (DA1E) mAb IgG XP® Isotype Control (CUT&RUN) #66362, using this CUT&RUN Assay Kit (left panel) or the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005 (right panel). The enriched DNA was quantified by real-time PCR using SimpleChIP® Human GAPDH Exon 1 Primers #5516 and SimpleChIP® Human α Satellite Repeat Primers #4486. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin, which is equivalent to one.
Figure 8. CUT&RUN and ChIP assays were performed with HCT 116 cells and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751, using this CUT&RUN Assay Kit or the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA Libraries were prepared using DNA Library Prep Kit for Illumina® (ChIP-seq, CUT&RUN) #56795. The upper panel compares enrichment of H3K4me3 across chromosome 12, while the lower panel compares enrichment at the GAPDH gene, a known target of H3K4me3. The input tracks are from the CUT&RUN input sample.
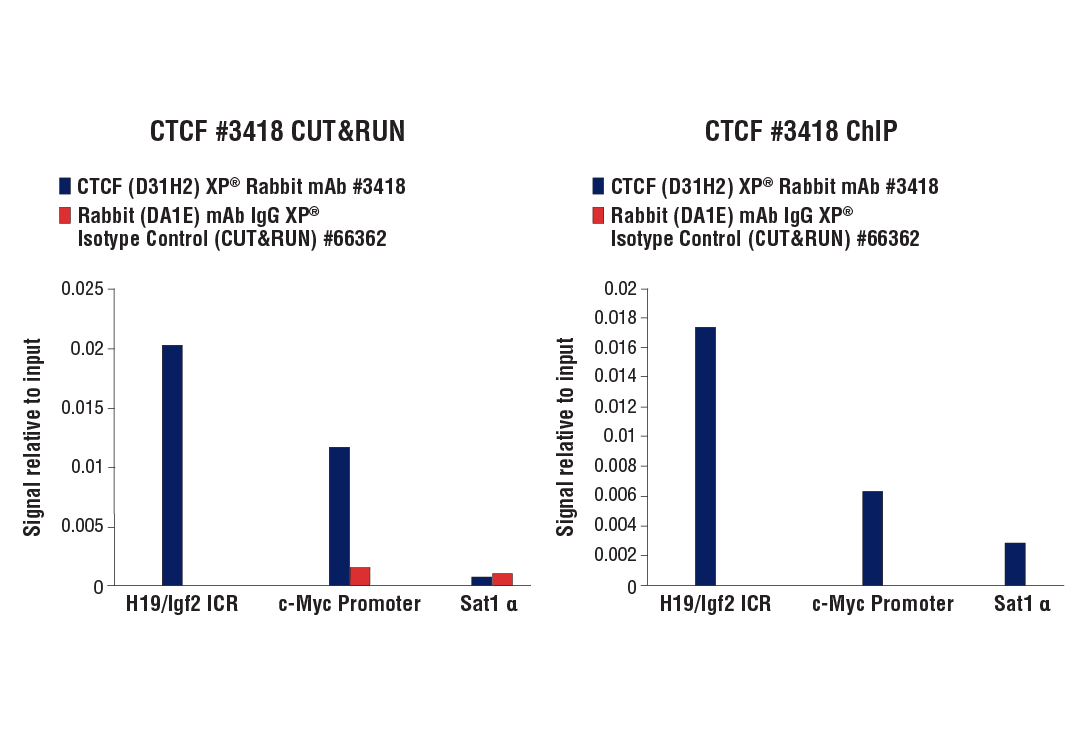
Figure 9. CUT&RUN and ChIP assays were performed with HCT 116 cells and CTCF (D31H2) XP® Rabbit mAb #3418 or Rabbit (DA1E) mAb IgG XP® Isotype Control (CUT&RUN) #66362, using this CUT&RUN Assay Kit (left panel) or the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005 (right panel). The enriched DNA was quantified by real-time PCR using human c-Myc promoter primers, SimpleChIP® Human H19/Igf2 ICR Primers #5172, and SimpleChIP® Human α Satellite Repeat Primers #4486. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin, which is equivalent to one.
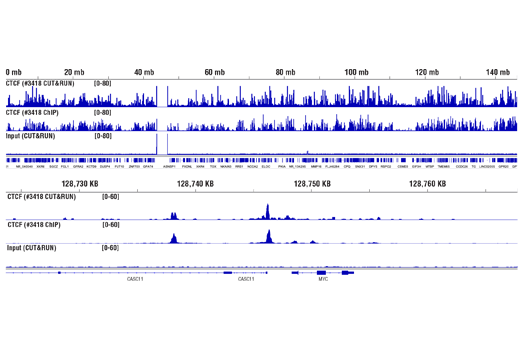
Figure 10. CUT&RUN and ChIP assays were performed with HCT 116 cells and CTCF (D31H2) XP® Rabbit mAb #3418, using this CUT&RUN Assay Kit or the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA Libraries were prepared using DNA Library Prep Kit for Illumina® (ChIP-seq, CUT&RUN) #56795. The upper panel compares enrichment of CTCF across chromosome 8, while the lower panel compares enrichment at the MYC gene, a known target of CTCF. The input tracks are from the CUT&RUN input sample.
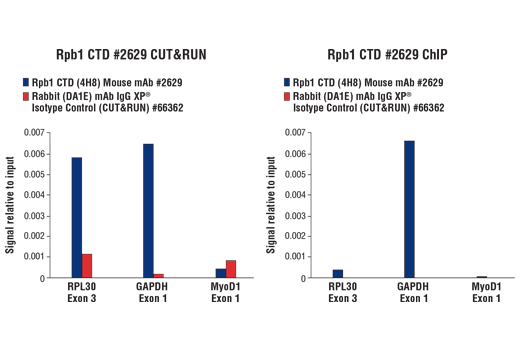
Figure 11. CUT&RUN and ChIP assays were performed with HeLa cells and Rpb1 CTD (4H8) Mouse mAb #2629 or Rabbit (DA1E) mAb IgG XP® Isotype Control (CUT&RUN) #66362, using this CUT&RUN Assay Kit (left panel) or the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005 (right panel). The enriched DNA was quantified by real-time PCR using SimpleChIP® Human RPL30 Exon 3 Primers #7014, SimpleChIP® Human GAPDH Exon 1 Primers #5516, and SimpleChIP® Human MyoD1 Exon 1 Primers #4490. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin, which is equivalent to one.