 全部商品分类
全部商品分类



 下载产品说明书
下载产品说明书 下载SDS
下载SDS 用小程序,查商品更便捷
用小程序,查商品更便捷


 收藏
收藏
 对比
对比 咨询
咨询The CUT&RUN pAG-MNase and Spike-In DNA kit provides enough enzyme and spike-in DNA to support 50 CUT&RUN assays. The pAG-MNase Enzyme #57813 is a fusion of Protein A and Protein G to Micrococcal Nuclease, and is recombinantly produced in E. coli. The pAG-MNase is compatible with multiple species of antibodies, including both rabbit and mouse. The Sample Normalization Spike-In DNA (10 pg/µl) #29987 is fragmented genomic DNA from the yeast S. cerevisiae that can be added to a CUT&RUN reaction to facilitate normalization between samples during NG-seq analysis.

Product Usage Information
For the pAG-MNase Enzyme, after cell permeabilization and primary antibody binding, resuspend cells in 50 μl of digitonin buffer containing 1.5 μl of pAG-MNase Enzyme (33X dilution). Incubate cell samples with rotation at 4°C for 1 hour, wash cells with digitonin buffer, and then perform the chromatin digestion. Sample Normalization Spike-In DNA can be added directly to the digestion stop buffer. For sample normalization with NG-seq, add 5 μl (50 pg) of Sample Normalization Spike-In DNA to each reaction. When using 100,000 cells or 1mg of tissue per reaction this ensures that the normalization reads are around 0.5% of the total sequencing reads. If more or less than 100,000 cells or 1mg of tissue are used per reaction, proportionally scale the volume of Sample Normalization Spike-In DNA up or down to adjust normalization reads to around 0.5% of total reads. When performing sample normalization, be sure to map the CUT&RUN sequencing data for all samples to both the test reference genome (e.g. human) and the sample normalization genome (S. cerevisiae).



pAG-MNase Enzyme is supplied in 10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1mM EDTA, 0.1mM PMSF and 50% glycerol. Do not aliquot the product. Sample Normalization Spike-In DNA is supplied in 10 mM Tris-HCl (pH 8.0). Store both products at –20°C. These products are stable for at least 12 months.
参考图片
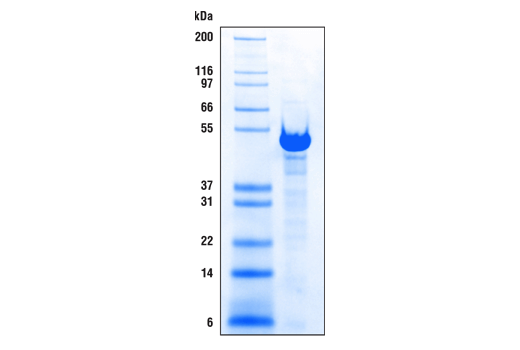
pAG-MNase was expressed and purified from E. coli. Purified enzyme was resolved on an SDA-PAGE gel and stained with Coomassie blue. The molecular weight of the protein standards is indicated.
CUT&RUN assays were performed with HCT 116 cells and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751. DNA Libraries were prepared using SimpleChIP® ChIP-seq DNA Library Prep Kit for Illumina® #56795. The size of the DNA fragments in the library were analyzed using an Agilent Bioanalyzer®. The adaptor and barcode sequences added to the library during construction account for 140 bp in fragment length. As shown, excised DNA is highly enriched for mononucleosomes (peak around 300 bp).
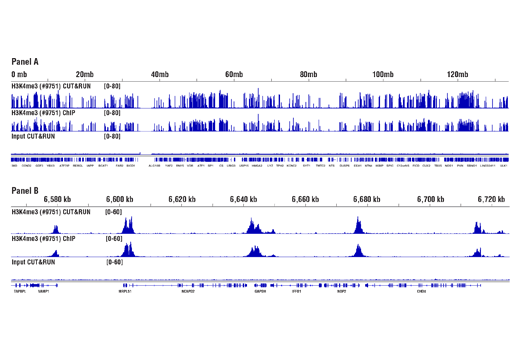
CUT&RUN and ChIP assays were performed with HCT 116 cells and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751. DNA Libraries were prepared using SimpleChIP® ChIP-seq DNA Library Prep Kit for Illumina® #56795. Panel A compares enrichment of H3K4me3 across chromosome 12 (upper), while Panel B compares enrichment at the GAPDH gene (lower), a known target of H3K4me3. The input tracks are from the CUT&RUN input sample.
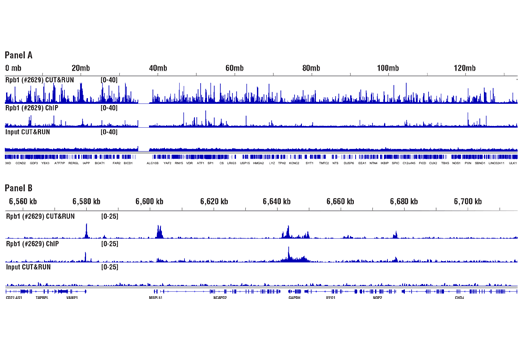
CUT&RUN and ChIP assays were performed with HeLa cells and Rpb1 CTD (4H8) Mouse mAb #2629. DNA Libraries were prepared using SimpleChIP® ChIP-seq DNA Library Prep Kit for Illumina® #56795. Panel A compares enrichment of Rpb1 across chromosome 12 (upper), while Panel B compares enrichment at the GAPDH gene (lower), a known target of Rbp1. The input tracks are from the CUT&RUN input sample.
CUT&RUN and ChIP assays were performed with HCT 116 cells and CTCF (D31H2) XP® Rabbit mAb #3418. DNA Libraries were prepared using SimpleChIP® ChIP-seq DNA Library Prep Kit for Illumina® #56795. Panel A compares enrichment of CTCF across chromosome 8 (upper), while Panel B compares enrichment at the MYC gene (lower), a known target of CTCF. The input tracks are from the CUT&RUN input sample.