 全部商品分类
全部商品分类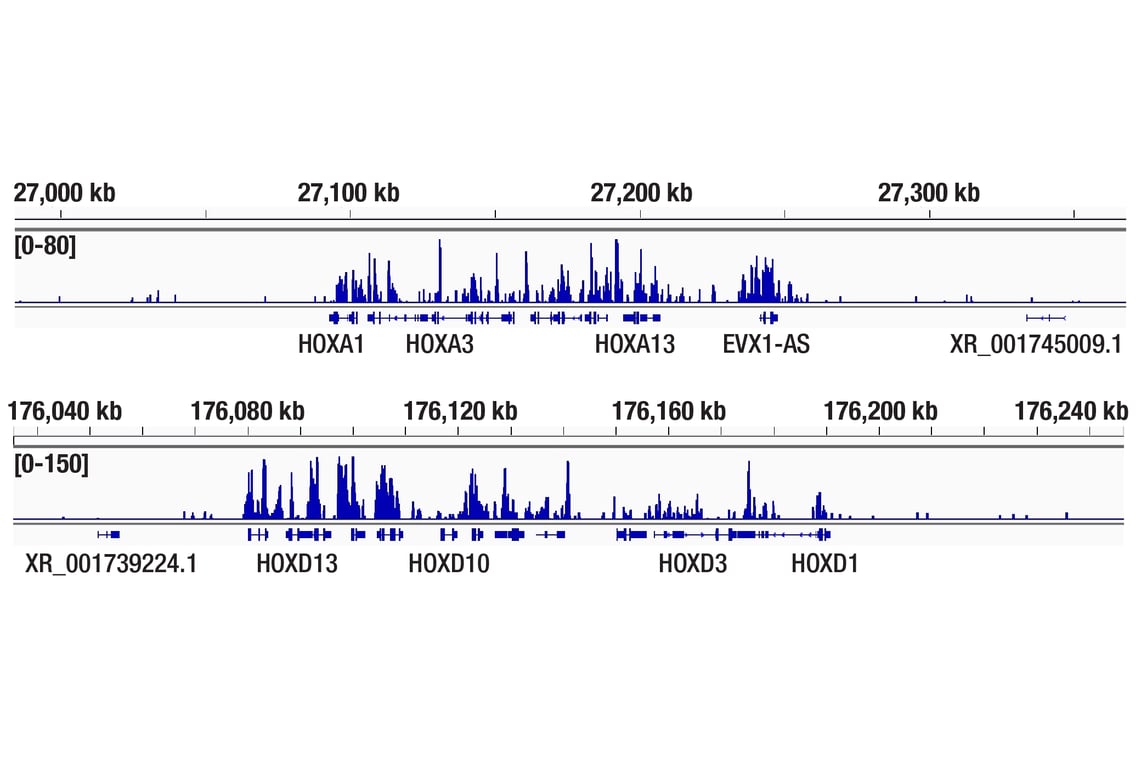



 下载产品说明书
下载产品说明书 下载COA
下载COA 下载SDS
下载SDS 用小程序,查商品更便捷
用小程序,查商品更便捷


 收藏
收藏
 对比
对比 咨询
咨询The CUT&Tag pAG-Tn5 (Loaded) enzyme provides enough enzyme to support 50 CUT&Tag assays. It is a fusion of Protein A and Protein G to Tn5, and is recombinantly produced in E. coli. This pAG-Tn5 has been loaded with the adaptor oligo that is compatible with NG-Sequencing for Illumina systems, so the genomic DNA fragments tagmented by CUT&Tag pAG-Tn5 (Loaded) are ready for PCR amplification and NG-seq. This enyzme is compatible with multiple species of antibodies, including both rabbit and mouse. This enzyme is validated using CUT&Tag Assay Kit #77552.

Product Usage Information
Please refer to CUT&Tag Assay Kit #77552 for detailed use of this enzyme in the CUT&Tag assay. After cell permeabilization and primary and secondary antibody binding, resuspend cells in 50 μL of High Salt Digitonin Buffer containing 2 μL of pAG-Tn5 Enzyme (1:25 dilution). Incubate cell samples at room temperature for 1 hour, wash cells with High Salt Digitonin Buffer, and then perform the chromatin tagmentation.

Specificity/Sensitivity
Species Reactivity:
All Species Expected



Supplied in 22 mM HEPES pH 7.4, 44 mM NaCl, 44 μM EDTA, 0.4 mM DTT, 0.04% Triton X-100, and 50% glycerol. Store at –20°C and do not aliquot. This product is stable for 6 months.
参考图片
Figure 3. CUT&Tag was performed with NCCIT cells and JARID2 (D6M9X) Rabbit mAb #13594, using CUT&Tag pAG-Tn5 (Loaded) and CUT&Tag Assay Kit #77552. The DNA library was prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figure shows binding across HOXA (upper) and HOXD (lower), known target genes of JARID2.
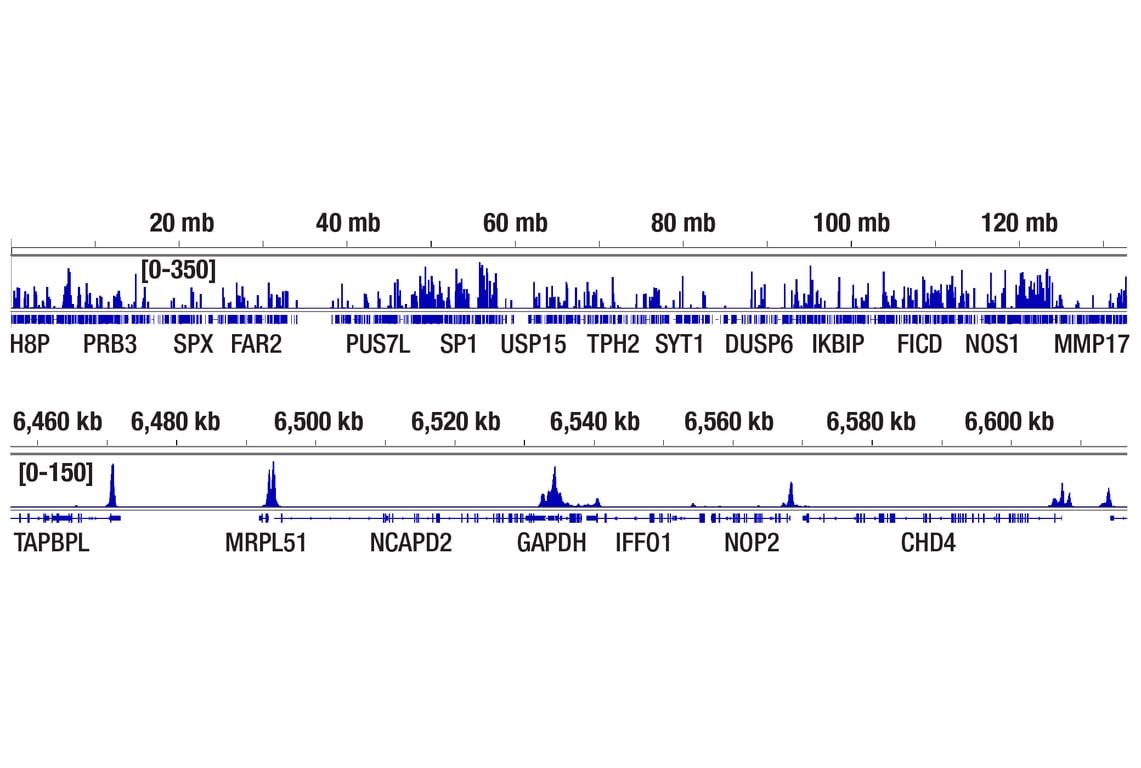
Figure 1. CUT&Tag was performed with HCT 116 cells and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751, using CUT&Tag pAG-Tn5 (Loaded) and CUT&Tag Assay Kit #77552. The DNA library was prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figure shows binding across chromosome 12 (upper), including GAPDH (lower), a known target gene of H3K4me3.
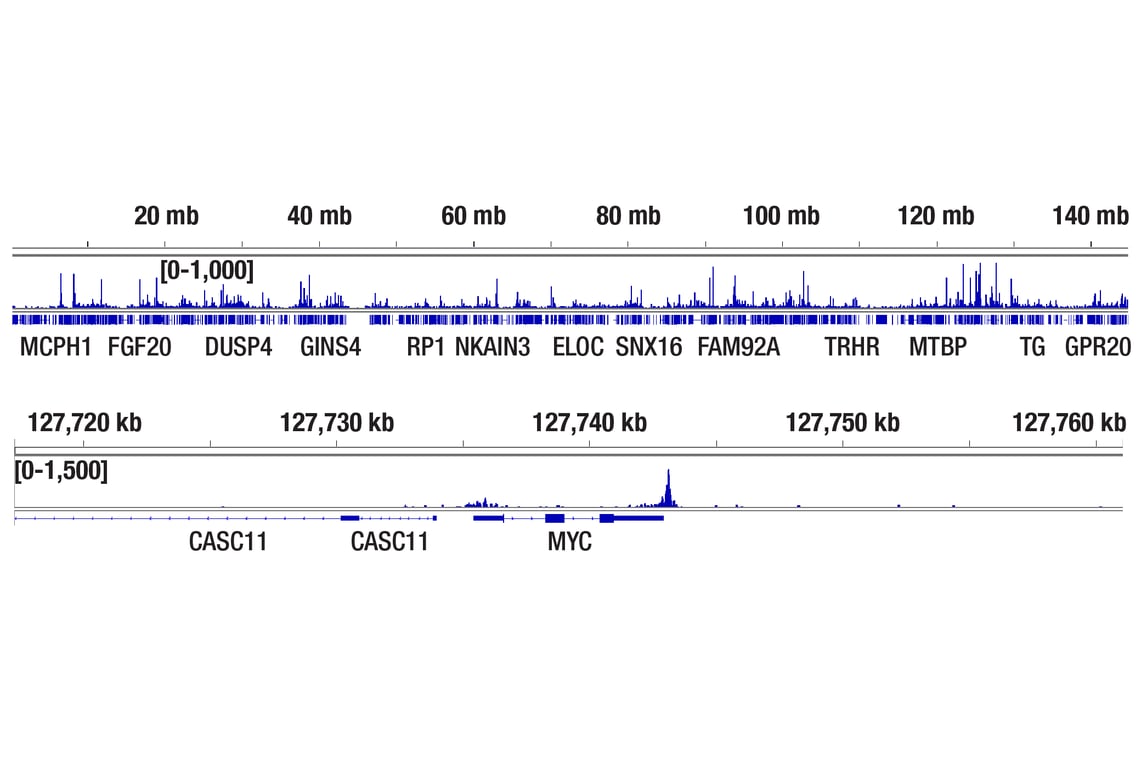
Figure 2. CUT&Tag was performed with HCT 116 cells and TCF4/TCF7L2 (C48H11) Rabbit mAb #2569, using CUT&Tag pAG-Tn5 (Loaded) and CUT&Tag Assay Kit #77552. The DNA library was prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figure shows binding across chromosome 8 (upper), including MYC (lower), a known target gene of TCF4.