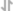 全部商品分类
全部商品分类


Next generation sequencing (NG-seq) is a high throughput method that can be used downstream of chromatin immunoprecipitation (ChIP) and Cleavage Under Targets and Release Using Nuclease (CUT&RUN) assays to identify and quantify target DNA enrichment across the entire genome. The DNA Library Prep Kit for Illumina Systems (ChIP-seq, CUT&RUN) contains all of the enzymes and buffers necessary to generate high quality DNA sequencing libraries from ChIP or CUT&RUN DNA for next-generation sequencing on the Illumina Systems platform. The fast, user-friendly workflow minimizes hands-on time needed for generation and purification of DNA libraries. This product must be used in combination with Multiplex Oligos for Illumina Systems (Single Index Primers) (ChIP-seq, CUT&RUN) #29580 or Multiplex Oligos for Illumina Systems (Dual Index Primers) (ChIP-seq, CUT&RUN) #47538.
This product provides sufficient amounts of reagents for 24 reactions and is compatible with both enzymatic- or sonication-fragmented, ChIP-enriched DNA. Distinct protocols are provied for DNA library preparation from ChIP and CUT&RUN DNA. This product is compatible with SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) #9003, SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005, SimpleChIP® Plus Sonication Chromatin IP Kit #56383, and CUT&RUN Assay Kit #86652. This product is not compatible with SimpleChIP® Enzymatic Chromatin IP Kit (Agarose Beads) #9002 and SimpleChIP® Plus Enzymatic Chromatin IP Kit (Agarose Beads) #9004 because agarose beads are blocked with sonicated salmon sperm DNA, which will contaminate DNA library preps and NG-seq.



Specificity/Sensitivity
Species Reactivity:
All Species Expected



Store all components at -20ºC. This product is stable for 12 months if stored properly.
参考图片
Figure 8. Agilent Bioanalyzer System profiles of DNA libraries prepared from different starting amounts of CUT&RUN DNA generated using the Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751. DNA libraries were prepared from 1 ng (A), 0.5 ng (B), 0.3 ng (C), and 0.1 ng (D) of enriched CUT&RUN DNA. As shown, each DNA library preparation shows a similar DNA fragment size.
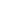
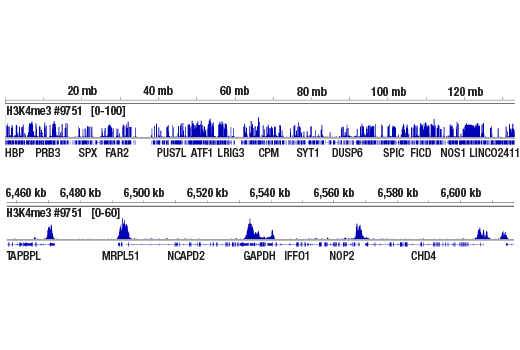
Figure 1. Chromatin immunoprecipitations were performed with cross-linked chromatin from 4 x 106 HCT 116 cells and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751, using SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA Libraries were prepared from 50 ng, 5 ng, or 0.5 ng enriched ChIP DNA or 50 ng Input DNA using SimpleChIP® ChIP-seq Multiplex Oligos for Illumina (Dual Index Primers) #47538, pooled into one sample, and sequenced on an Illumina Next-Seq platform. The figure shows binding across GAPDH, a known target gene of H3K4me3. For additional ChIP-seq tracks, please download the product datasheet.
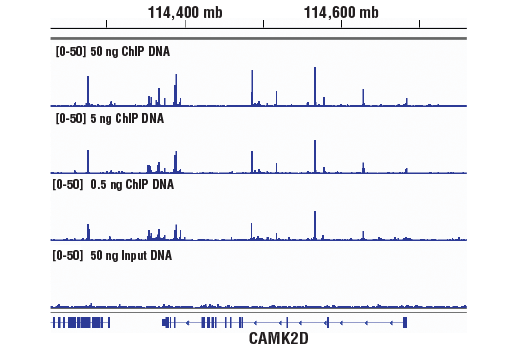
Figure 2. Chromatin immunoprecipitations were performed with cross-linked chromatin from 4 x 106 HCT 116 cells and TCF4/TCF7L2 (C48H11) Rabbit mAb #2569, using SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA Libraries were prepared from 50 ng, 5 ng, or 0.5 ng enriched ChIP DNA or 50 ng Input DNA using SimpleChIP® ChIP-seq Multiplex Oligos for Illumina (Dual Index Primers) #47538, pooled into one sample, and sequenced on an Illumina Next-Seq platform. The figure shows binding across CaMK2D, a known target gene of TCF4/TCF7L2. For additional ChIP-seq tracks, please download the product datasheet.
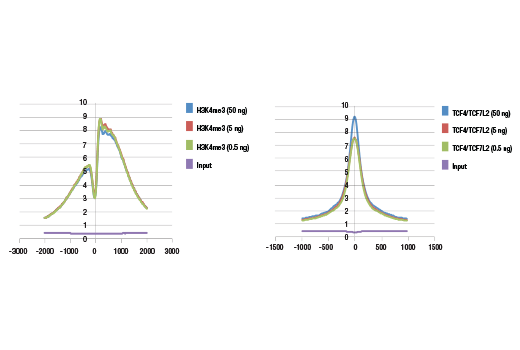
Figure 3. Metagene analysis of ChIP-seq data generated using different amounts of starting ChIP DNA. Analyses of both Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 (left) and TCF4/TCF7L2 (C48H11) Rabbit mAb #2569 (right) ChIP-seq data show that the signal-to-noise ratio for peaks identified across entire genome are similar for all three starting amounts of ChIP DNA.
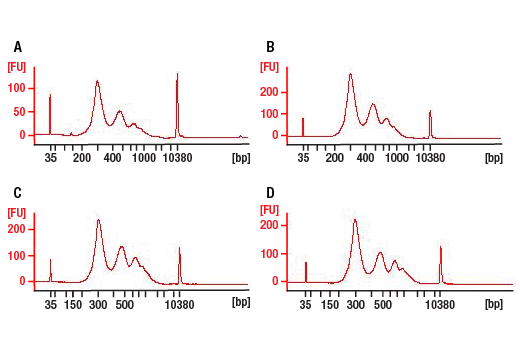
Figure 4. Agilent Bioanalyzer System profiles of DNA libraries prepared from different starting amounts of ChIP DNA generated using the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA libraries were prepared from 50 ng (A), 5 ng (B), and 0.5 ng (C) of enriched ChIP DNA and 50 ng (D) input DNA. As shown, each DNA library preparation shows a similar DNA fragment size (expected range 300 bp to 900 bp).
Figure 5. CUT&RUN NGS data generated using Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 and 3.6 ng of CUT&RUN DNA, as described in Table 2. The figure shows binding across chromosome 12 (upper), including GAPDH (lower), a known target gene of H3K4me3.
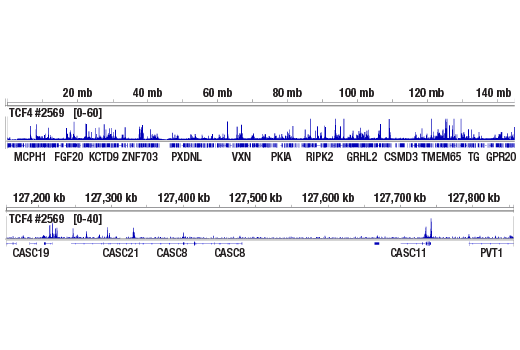
Figure 6. CUT&RUN NGS data generated using TCF4/TCF7L2 (C48H11) Rabbit mAb #2569 and 0.7 ng of CUT&RUN DNA, as described in Table 2. The figure shows binding across chromosome 8 (upper), including MYC (lower), a known target gene of TCF4/TCF7L2.
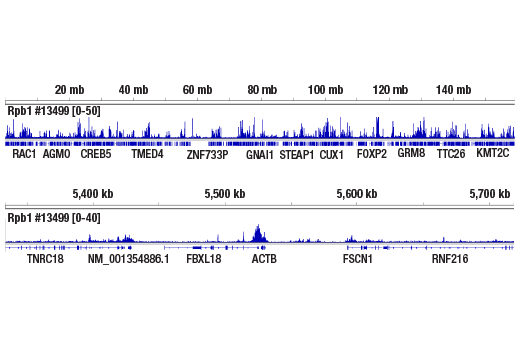
Figure 7. CUT&RUN NGS data generated using Phospho-Rpb1 CTD (Ser2) (E1Z3G) Rabbit mAb #13499 and 3.1 ng of CUT&RUN DNA, as described in Table 2. The figure shows binding across chromosome 7 (upper), including ACTB (lower), a known target gene of Phospho-Rpb1.






 用小程序,查商品更便捷
用小程序,查商品更便捷