 全部商品分类
全部商品分类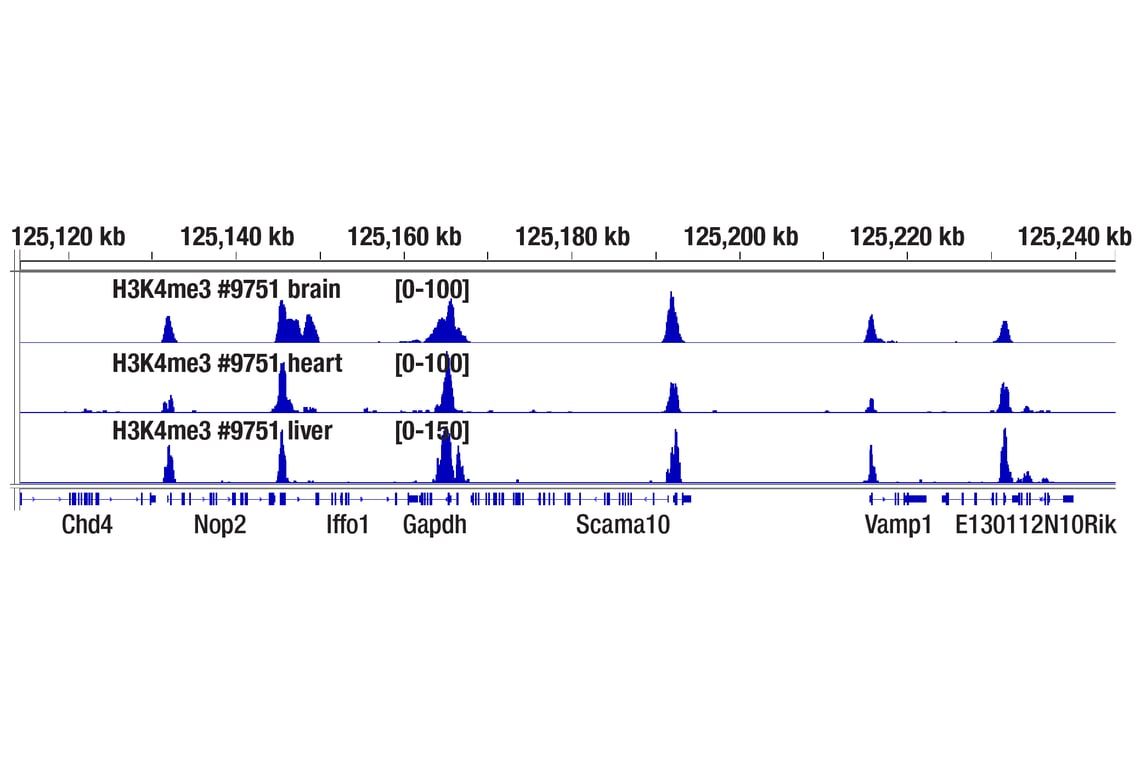



 下载产品说明书
下载产品说明书 下载SDS
下载SDS 用小程序,查商品更便捷
用小程序,查商品更便捷


 收藏
收藏
 对比
对比 咨询
咨询The CUT&Tag Assay Kit is designed to conveniently provide reagents needed to perform up to 24 reactions. The kit has been optimized to work in fresh or lightly fixed cells for all types of DNA-binding proteins, including histones, transcription factors, and cofactors. In addition, the kit has been optimized to work for histones in fresh or lightly fixed tissues. For analysis of transcription factors and cofactors in tissues, we recommend using the CUT&RUN Assay Kit #86652. If possible, we recommend using 100,000 cells or 1 mg of tissue per CUT&Tag reaction. If starting cell number is limited, this kit is validated to work with as few as 5,000 to 10,000 cells per reaction for histone modification targets and as few as 20,000 cells per reaction for transcription factors and cofactors. This kit is compatible with both whole cells and nuclei as starting material. We have not found that using nuclei generates a stronger signal or a higher signal-to-noise reaction compared to whole cells. A complete assay can be performed in as little as one day. The CUT&Tag Assay Kit also provides important controls to ensure a successful CUT&Tag experiment, including positive control Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 and negative controls Normal Rabbit IgG #2729 and Normal Mouse IgG #68860. The negative control Normal Rabbit IgG #2729 or Normal Mouse IgG #68860 is optional for the experiments, depending on the requirements of the peak calling algorithm used. This kit is compatible with downstream Next Generation sequencing (NG-seq) analysis, but not qPCR.



Specificity/Sensitivity



All components in this kit are stable for 6 months when stored at the recommended temperature.
参考图片
Figure 7. CUT&Tag assay was performed with 1 mg of fresh mouse brain, heart, or liver tissues (as indicated) and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751, using this CUT&Tag Assay Kit. DNA libraries were prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figure shows enrichment of H3K4me3 around its known target gene GAPDH.
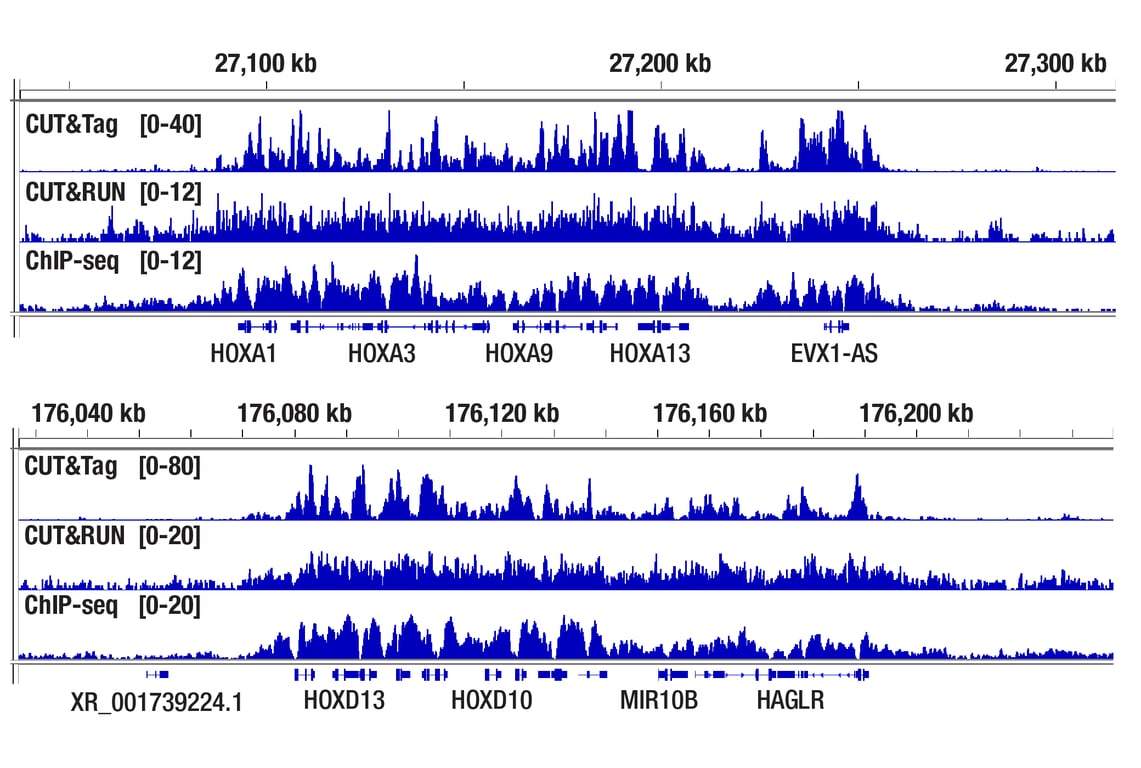
Figure 1. CUT&Tag, CUT&RUN, and ChIP-seq assays were performed with NCCIT cells and Tri-Methyl-Histone H3 (Lys27) (C36B11) Rabbit mAb #9733, using this CUT&Tag Assay Kit, the CUT&RUN Assay Kit #86652, or the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA libraries were prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415 for CUT&Tag samples and DNA Library Prep Kit for Illumina Systems (ChIP-seq, CUT&RUN) #56795 for ChIP-seq and CUT&RUN samples. The upper panel compares enrichment around HoxA genes, while the lower panel compares enrichment around HoxD genes, both are known target genes of H3K27me3.
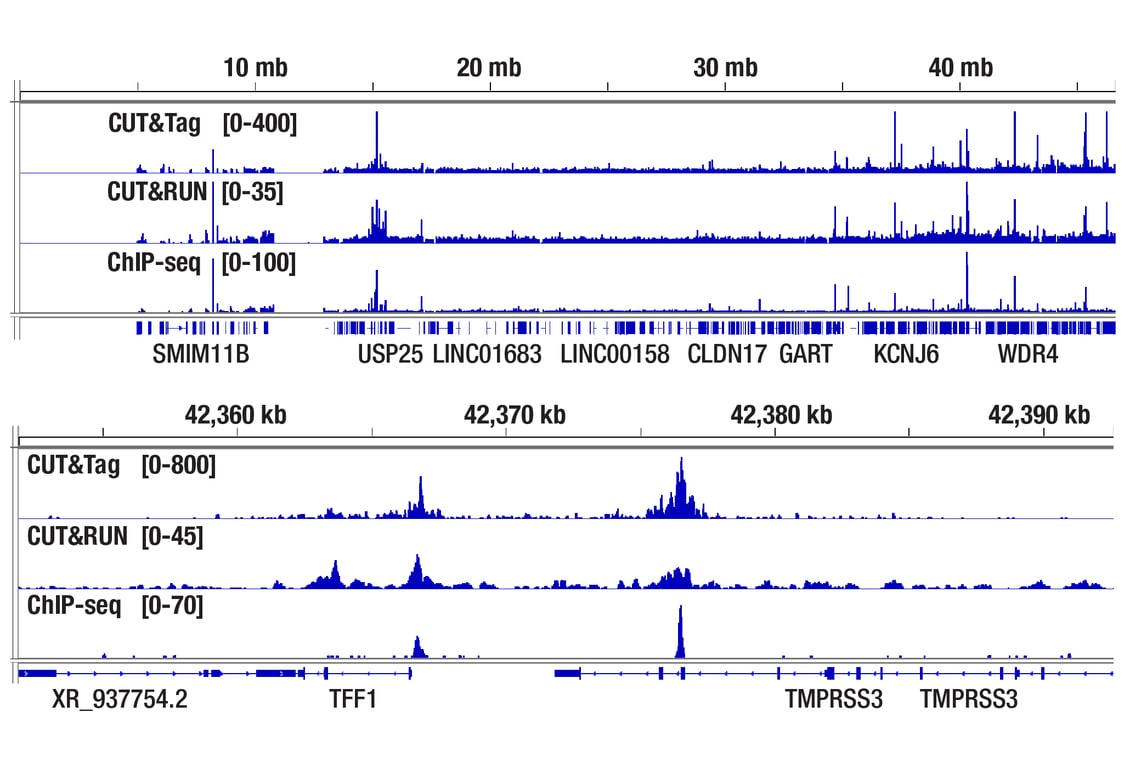
Figure 2. CUT&Tag, CUT&RUN, and ChIP-seq assays were performed with MCF7 cells grown in phenol red free medium and 5% charcoal stripped FBS for 4 d, then treated with β-estradiol (10 nM) for 45 min and Estrogen Receptor α (D8H8) Rabbit mAb #8644, using this CUT&Tag Assay Kit, the CUT&RUN Assay Kit #86652, or the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA libraries were prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415 for CUT&Tag samples and DNA Library Prep Kit for Illumina Systems (ChIP-seq, CUT&RUN) #56795 for ChIP-seq and CUT&RUN samples. The upper panel compares enrichment of Estrogen Receptor α across chromosome 21, while the lower panel compares enrichment around TFF1, a known target gene of Estrogen Receptor α.
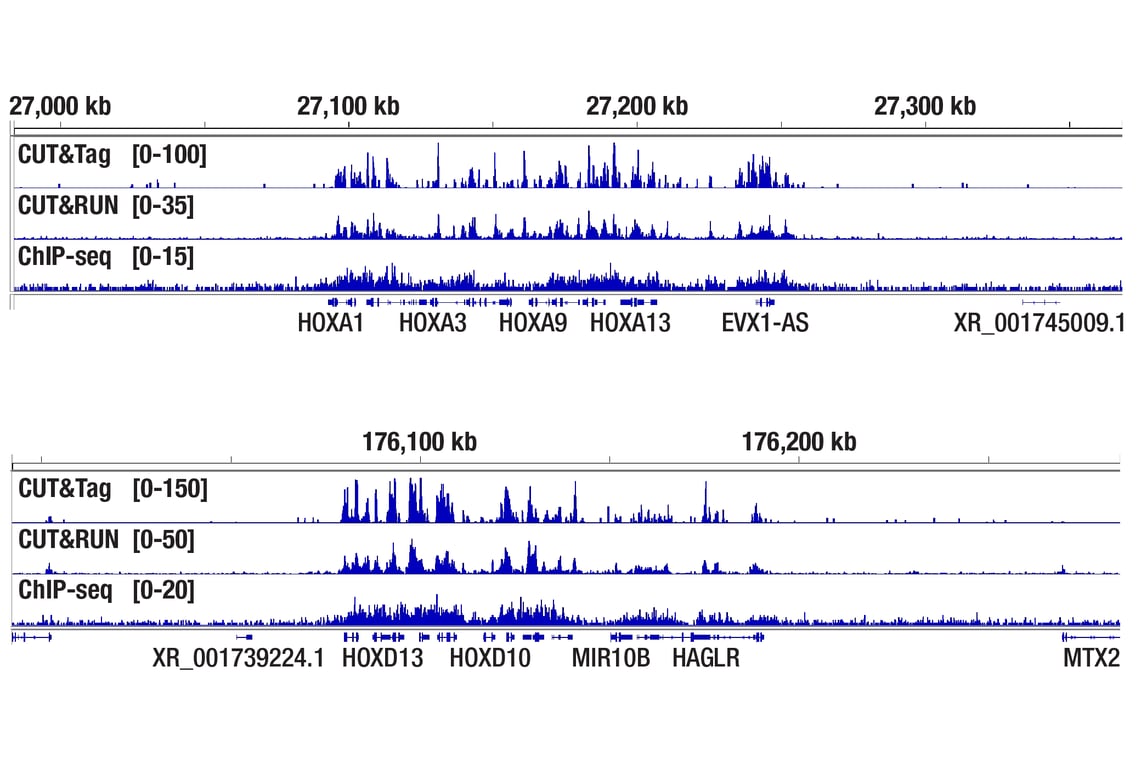
Figure 3. CUT&Tag, CUT&RUN, and ChIP-seq assays were performed with NCCIT cells and JARID2 (D6M9X) Rabbit mAb #13594, using this CUT&Tag Assay Kit, the CUT&RUN Assay Kit #86652, or the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA libraries were prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415 for CUT&Tag samples and DNA Library Prep Kit for Illumina Systems (ChIP-seq, CUT&RUN) #56795 for ChIP-seq and CUT&RUN samples. The upper panel compares enrichment around HoxA genes, while the lower panel compares enrichment around HoxD genes, both are known target genes of JARID2.
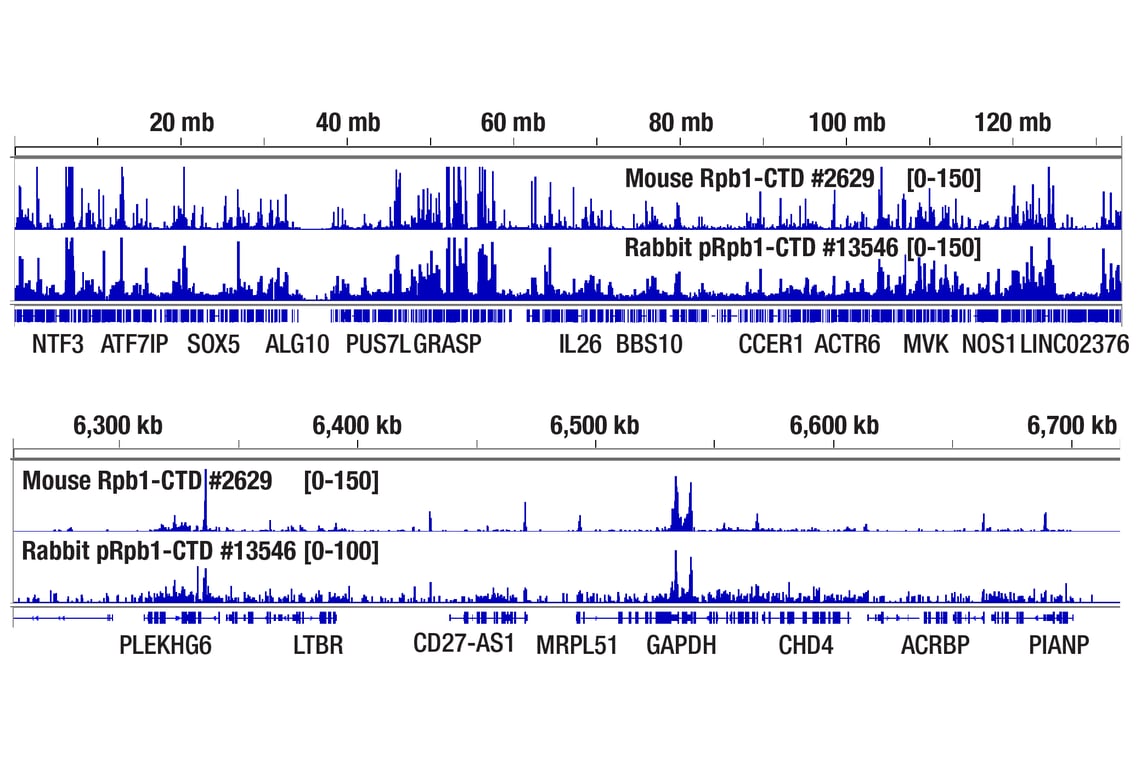
Figure 4. CUT&Tag assay was performed with HeLa cells and either Rpb1 CTD (4H8) Mouse mAb #2629 or Phospho-Rpb1 CTD (Ser2/Ser5) (D1G3K) Rabbit mAb #13546, using this CUT&Tag Assay Kit. DNA libraries were prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The upper panel compares enrichment of Rpb1 across chromosome 12, while the lower panel compares enrichment around GAPDH, a known target gene of Rpb1.
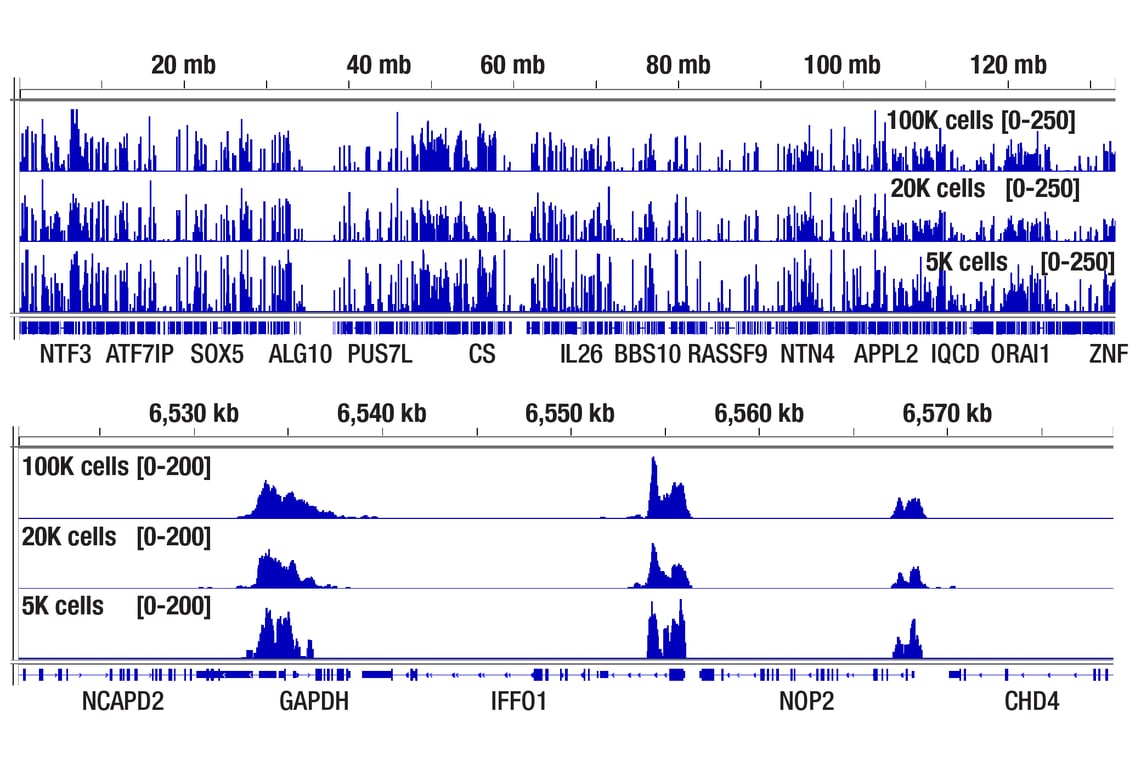
Figure 5. CUT&Tag assay was performed with 100,000, 20,000, or 5,000 NCCIT cells (as indicated) and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751, using this CUT&Tag Assay Kit. DNA libraries were prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The upper panel compares enrichment of H3K4me3 across chromosome 12, while the lower panel compares enrichment around GAPDH, a known target gene of H3K4me3.
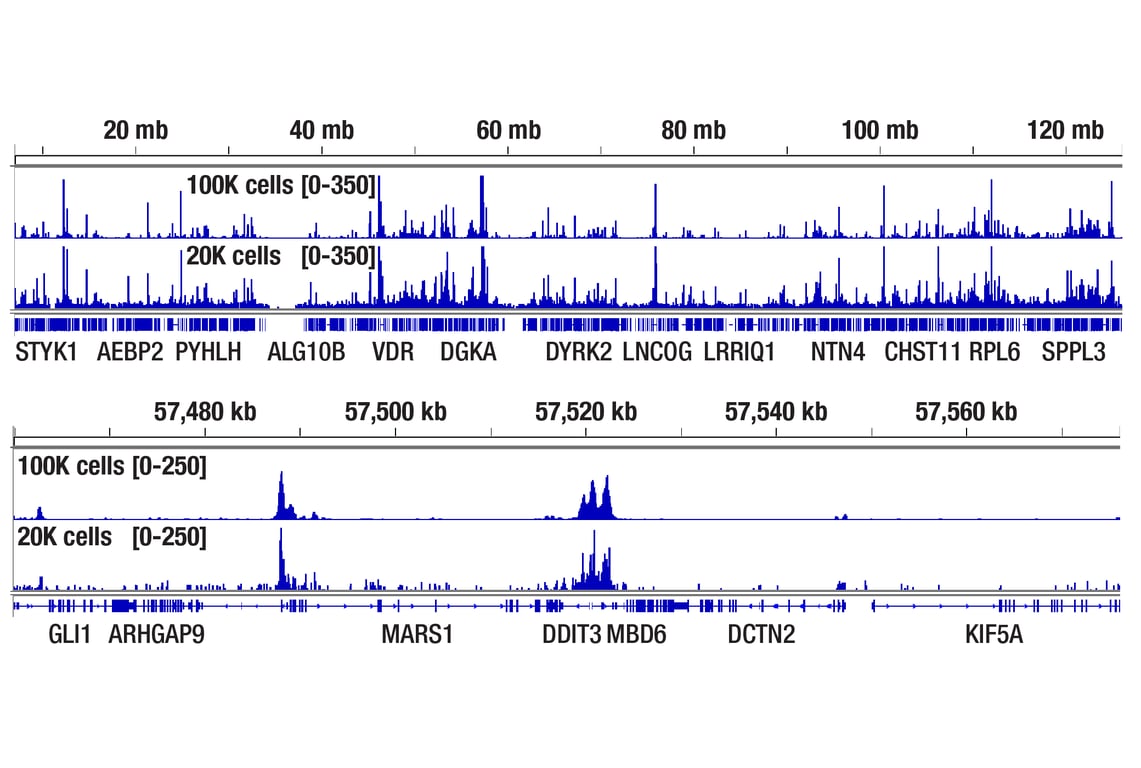
Figure 6. CUT&Tag assay was performed with 100,000 or 20,000 Hep G2 cells treated with Thapsigargin #12758 (300 nM) for 4h (as indicated) and ATF-4 (D4B8) Rabbit mAb #11815, using this CUT&Tag Assay Kit. DNA libraries were prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The upper panel compares enrichment of ATF-4 across chromosome 12, while the lower panel compares enrichment around DDIT3/CHOP, a known target gene of ATF-4.