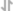 全部商品分类
全部商品分类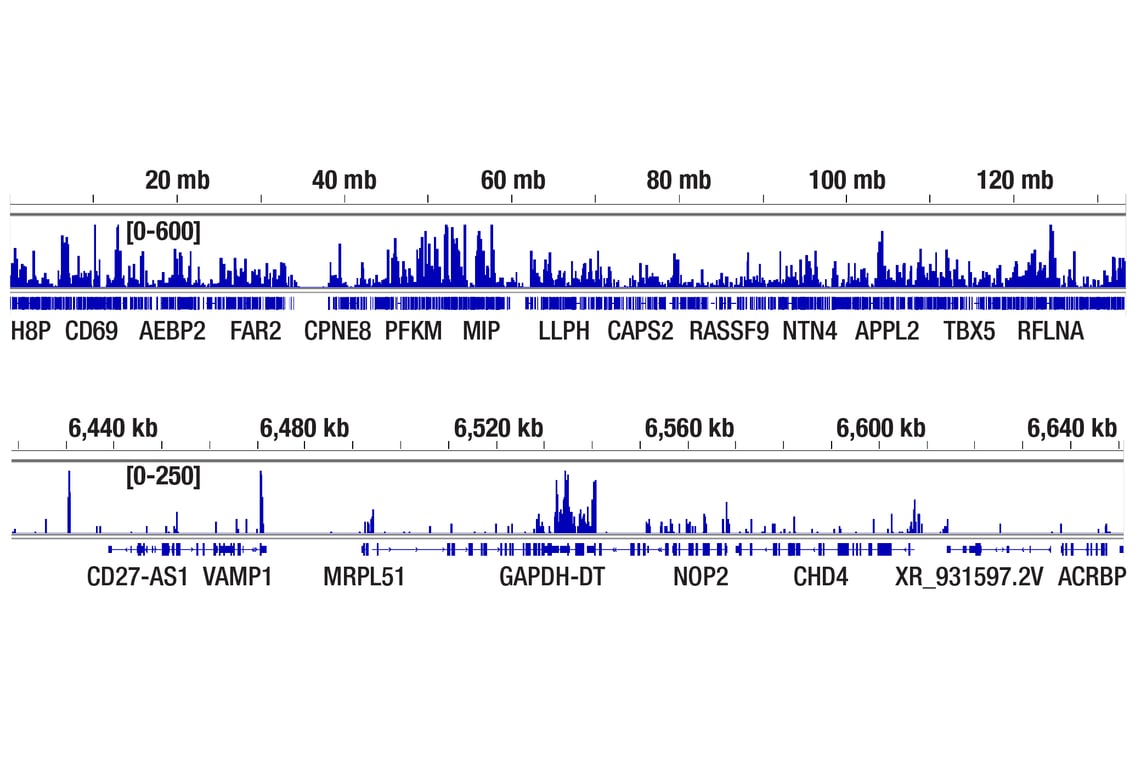



Monoclonal antibody is produced by immunizing animals with a synthetic phosphopeptide containing 10 heptapeptide repeats [Tyr1, Ser2, Pro3, Thr4, Ser5, Pro6, Ser7] in which Ser5 is phosphorylated.


Product Usage Information
For optimal ChIP and ChIP-seq results, use 10 μl of antibody and 10 μg of chromatin (approximately 4 x 106 cells) per IP. This antibody has been validated using SimpleChIP® Enzymatic Chromatin IP Kits. The CUT&RUN dilution was determined using CUT&RUN Assay Kit #86652. The CUT&Tag dilution was determined using CUT&Tag Assay Kit #77552.
| Application | Dilution |
|---|---|
| Western Blotting | 1:1000 |
| Simple Western™ | 1:50 - 1:250 |
| Immunoprecipitation | 1:50 |
| Chromatin IP | 1:50 |
| Chromatin IP-seq | 1:50 |
| CUT&RUN | 1:50 |
| CUT&Tag | 1:50 |







Specificity/Sensitivity
Species Reactivity:
Human, Mouse, Rat, Monkey




Supplied in 10 mM sodium HEPES (pH 7.5), 150 mM NaCl, 100 µg/ml BSA, 50% glycerol and less than 0.02% sodium azide. Store at –20°C. Do not aliquot the antibody.


参考图片
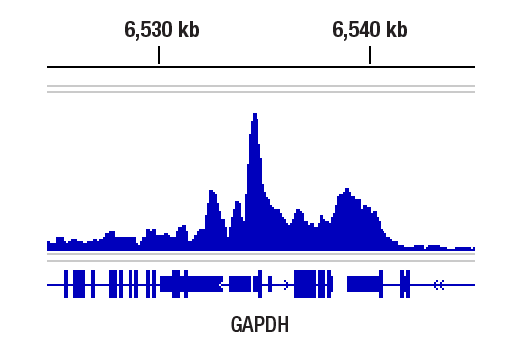
CUT&Tag was performed with HeLa cells and Rpb1 CTD (4H8) Mouse mAb, using CUT&Tag Assay Kit #77552. DNA library was prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figures show binding across chromosome 12 (upper), including GAPDH (lower), a known target gene of Rpb1 (see our ChIP-qPCR figure).
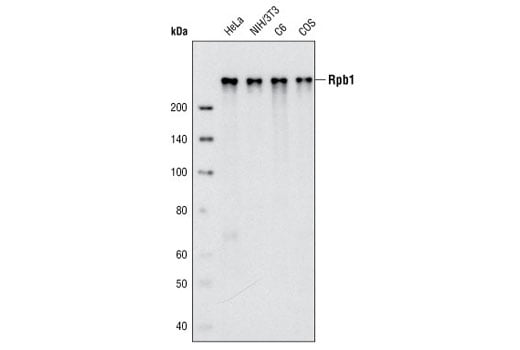
Western blot analysis of extracts from various cell lines using Rpb1 CTD (4H8) Mouse mAb.
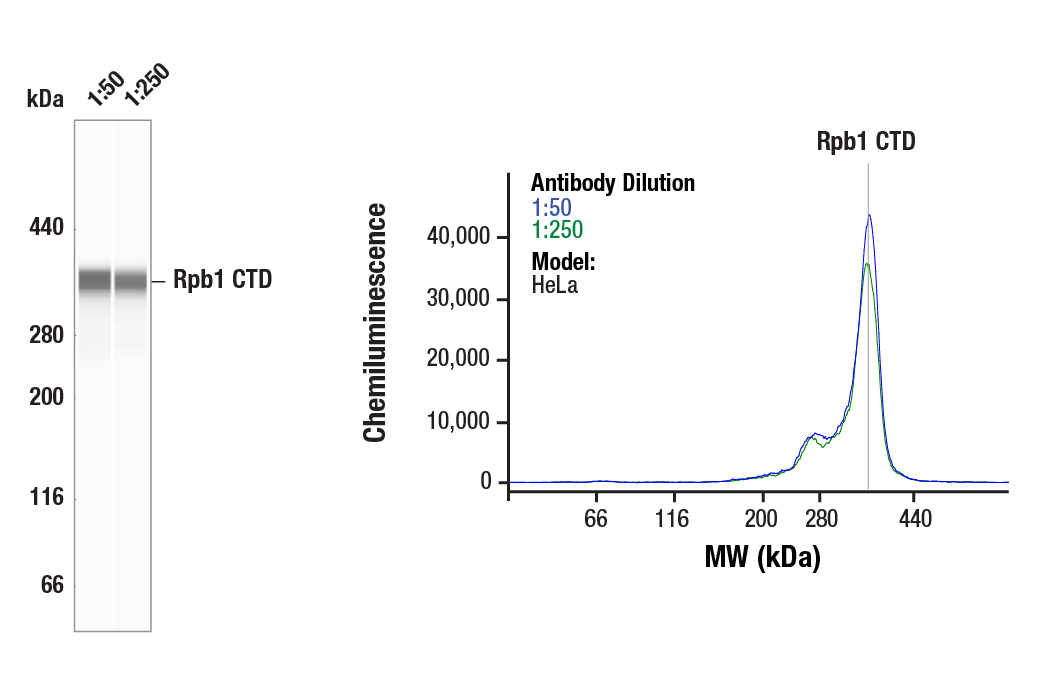
Simple Western™ analysis of lysates (0.1 mg/mL) from HeLa cells using Rpb1 CTD (4H8) Mouse mAb #2629. The virtual lane view (left) shows the target band (as indicated) at 1:50 and 1:250 dilutions of primary antibody. The corresponding electropherogram view (right) plots chemiluminescence by molecular weight along the capillary at 1:50 (blue line) and 1:250 (green line) dilutions of primary antibody. This experiment was performed under reducing conditions on the Jess™ Simple Western instrument from ProteinSimple, a BioTechne brand, using the 66-440 kDa separation module.
Chromatin immunoprecipitations were performed with cross-linked chromatin from HeLa cells and Rpb1 CTD (4H8) Mouse mAb, using SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The figure shows binding across GAPDH, a known target gene of Rpb1.
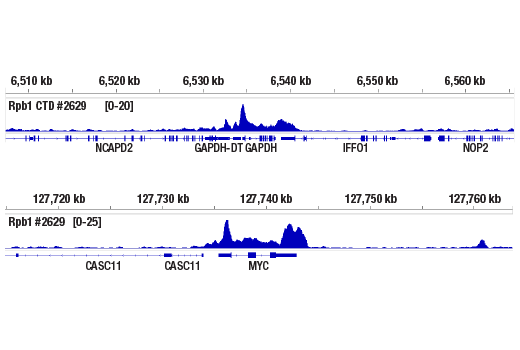
Chromatin immunoprecipitations were performed with cross-linked chromatin from HeLa cells and Rpb1 CTD (4H8) Mouse mA using, SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The figures show binding across GAPDH (upper) and Myc (lower), known target genes of Rpb1 (see additional figure containing ChIP-qPCR data).
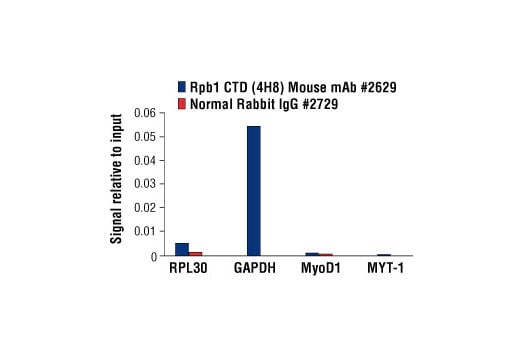
Chromatin immunoprecipitations were performed with cross-linked chromatin from HeLa cells and either Rpb1 CTD (4H8) Mouse mAb Normal Rabbit IgG #2729 using SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) #9003. The enriched DNA was quantified by real-time PCR using SimpleChIP® Human RPL30 Exon 3 Primers #7014, SimpleChIP® Human GAPDH Exon 1 Primers #5516, SimpleChIP® Human MyoD1 Exon 1 Primers #4490, and SimpleChIP® Human MYT-1 Exon 1 Primers #4493. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin, which is equivalent to one.
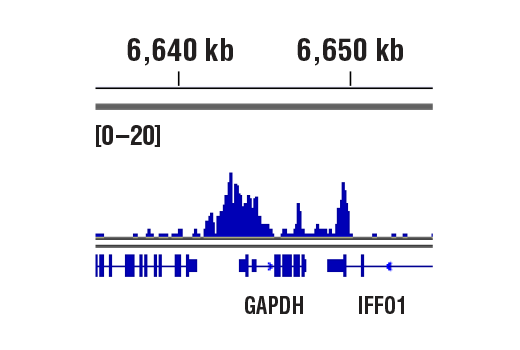
CUT&RUN was performed with HeLa cells and Rpb1 CTD (4H8) Mouse mAb, using CUT&RUN Assay Kit #86652. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The figure shows binding across GAPDH, a known target gene of Rpb1 (see additional figure containing CUT&RUN-qPCR data).
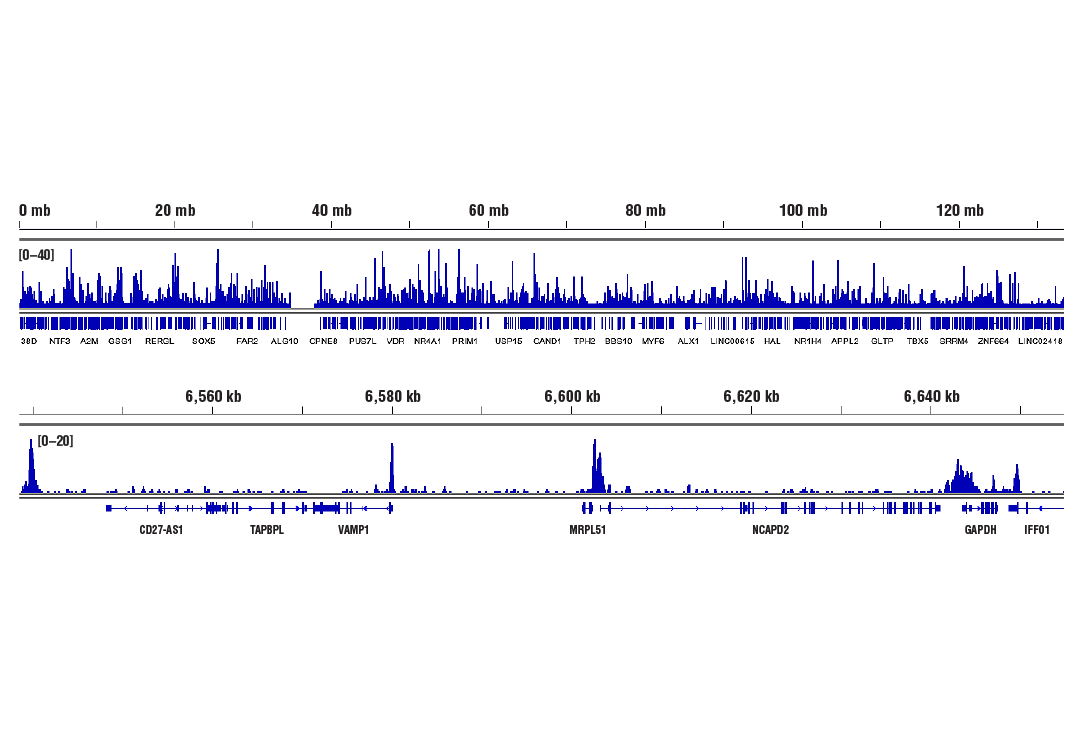
CUT&RUN was performed with HeLa cells and Rpb1 CTD (4H8) Mouse mAb, using CUT&RUN Assay Kit #86652. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The figures show binding across chromosome 12 (upper), including GAPDH (lower), a known target gene of Rpb1 (see additional figure containing CUT&RUN-qPCR data).
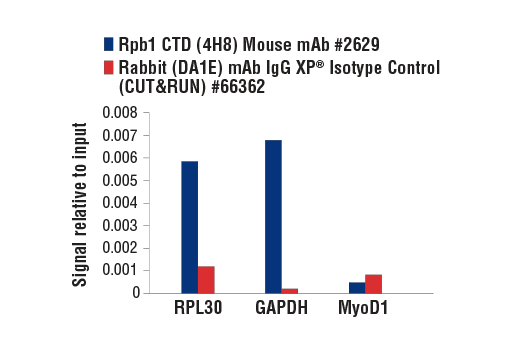
CUT&RUN was performed with HeLa cells and either Rpb1 CTD (4H8) Mouse mAb or Rabbit (DA1E) mAb IgG XP® Isotype Control (CUT&RUN) #66362, using CUT&RUN Assay Kit #86652. The enriched DNA was quantified by real-time PCR using SimpleChIP® Human RPL30 Exon 3 Primers #7014, SimpleChIP® Human GAPDH Exon 1 Primers #5516, and SimpleChIP® Human MyoD1 Exon 1 Primers #4490. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin, which is equivalent to one.
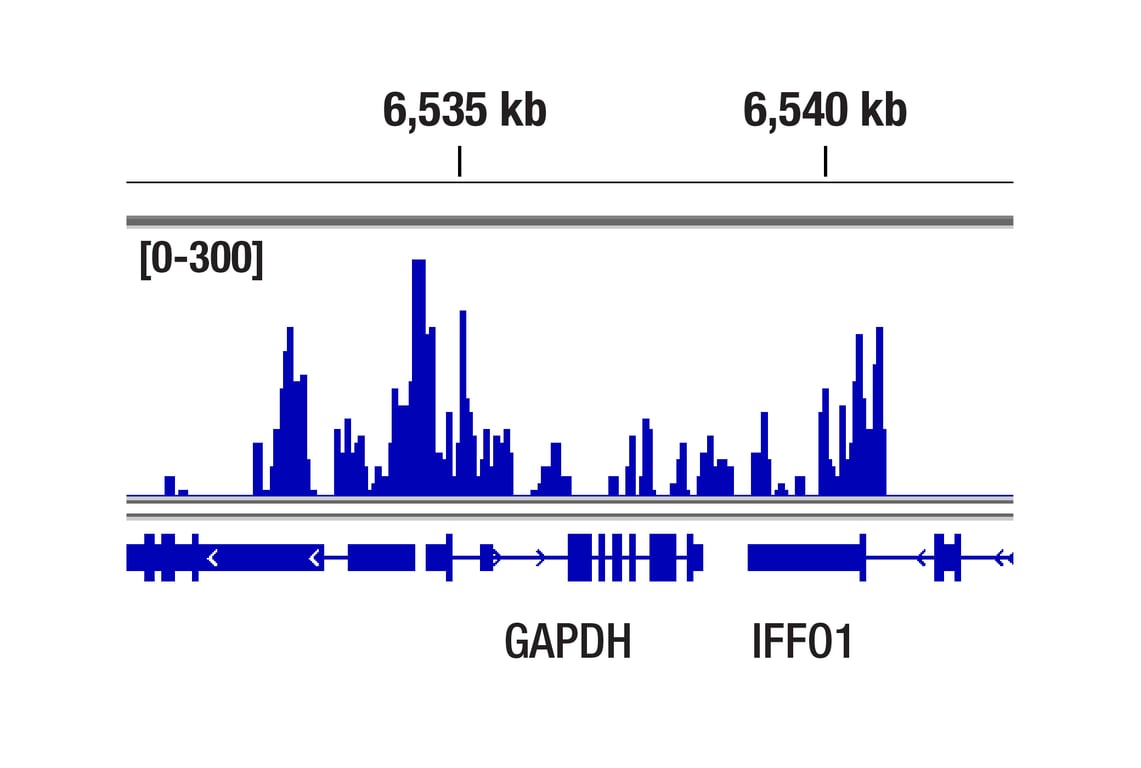
CUT&Tag was performed with HeLa cells and Rpb1 CTD (4H8) Mouse mAb, using CUT&Tag Assay Kit #77552. DNA library was prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figure shows binding across GAPDH, a known target gene of Rpb1 (see our ChIP-qPCR figure).





 用小程序,查商品更便捷
用小程序,查商品更便捷