 全部商品分类
全部商品分类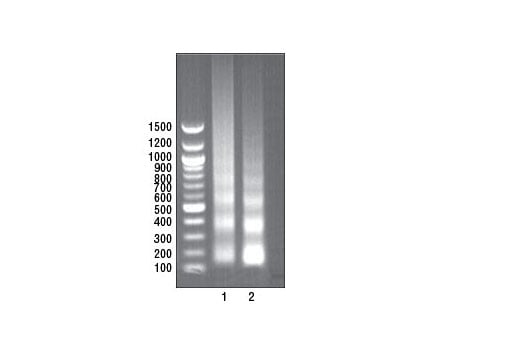


The SimpleChIP® Plus Enzymatic Chromatin IP Kit (Agarose Beads) #9004 contains the buffers and reagents necessary to perform up to 30 chromatin immunoprecipitations from cells or tissue samples, and is optimized for 4 X 106 cells or 25 mg of tissue per immunoprecipitation. A complete assay can be performed in as little as two days and can easily be scaled up or down for use with more or less cells or tissue sample.
Cells or tissue are fixed with formaldehyde and lysed, and chromatin is fragmented by partial digestion with Micrococcal Nuclease to obtain chromatin fragments of 1 to 5 nucleosomes. Enzymatic fragmentation of chromatin is much milder than sonication and eliminates problems resulting from variability in sonication power and emulsification of chromatin during sonication, which can result in incomplete fragmentation of chromatin or loss of antibody epitopes due to protein denaturation and degradation. Chromatin immunoprecipitations are performed using ChIP-validated antibodies and ChIP-Grade Protein G Agarose Beads. After reversal of protein-DNA cross-links, the DNA is purified using DNA purification spin columns, allowing for easy and efficient recovery of DNA and removal of protein contaminants without the need for phenol/chloroform extractions and ethanol precipitations. The enrichment of particular DNA sequences during immunoprecipitation can be analyzed by a variety of methods, including standard PCR and quantitative real-time PCR. Please note that this kit is not compatible with ChIP-seq because the ChIP-Grade Protein G Agarose Beads are blocked with sonicated salmon sperm DNA, which interferes with downstream sequencing.
The SimpleChIP® Plus Kit also provides important controls to ensure a successful ChIP experiment. The kit contains a positive control Histone H3 Antibody, a negative control Normal Rabbit IgG Antibody and primer sets for PCR detection of the human and mouse ribosomal protein L30 (RPL30) genes. Histone H3 is a core component of chromatin and is bound to most DNA sequences throughout the genome, including the RPL30 locus. Thus, the Histone H3 Antibody provides a universal positive control that should enrich for almost any locus examined.




Specificity/Sensitivity



Please store components at the temperatures indicated on the individual tube labels.
参考图片
FIGURE 3. Mouse brain or mouse liver were cross-linked and disaggregated into a single-cell suspension using a Dounce homogenizer or tissue disaggregator, respectively. The chromatin was prepared and digested, and chromatin immunoprecipitations were performed using the indicated ChIP-validated antibodies. DNA was purified and 10 μl was separated by electrophoresis on a 1% agarose gel and stained with ethidium bromide. The majority of chromatin from both brain (lane 1) and liver (lane 2) was digested to 1 to 5 nucleosomes in length (150 to 900 bp).
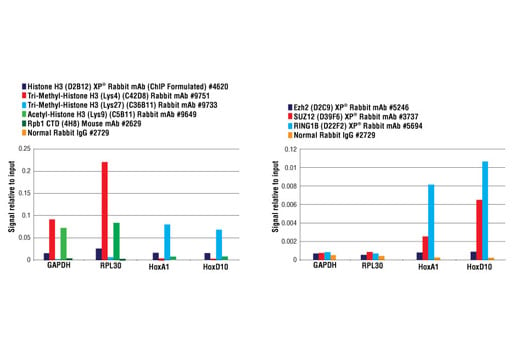
FIGURE 1. Mouse brain was cross-linked and disaggregated into a single-cell suspension using a Dounce homogenizer. The chromatin was prepared and digested, and chromatin immunoprecipitations were performed using the indicated ChIP-validated antibodies. Purified DNA was analyzed by quantitative real-time PCR using SimpleChIP® Mouse GAPDH Intron 2 Primers #8986, SimpleChIP® Mouse RPL30 Intron 2 Primers #7015, SimpleChIP® Mouse HoxA1 Promoter Primers #7341, and SimpleChIP® Mouse HoxD10 Exon 1 Primers #7429. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin (equivalent to 1).
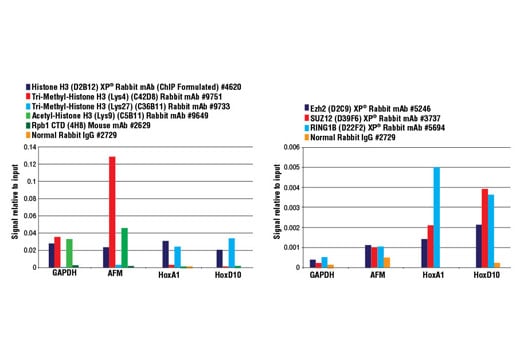
FIGURE 2. Mouse liver was cross-linked and disaggregated into a single-cell suspension using a tissue disaggregator. The chromatin was prepared and digested, and chromatin immunoprecipitations were performed using the indicated ChIP-validated antibodies. Purified DNA was analyzed by quantitative real-time PCR using SimpleChIP® Mouse GAPDH Intron 2 Primers #8986, SimpleChIP® Mouse AFM Intron 2 Primers #7269, SimpleChIP® Mouse HoxA1 Promoter Primers #7341, and SimpleChIP® Mouse HoxD10 Exon 1 Primers #7429. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin (equivalent to 1).






 用小程序,查商品更便捷
用小程序,查商品更便捷