 全部商品分类
全部商品分类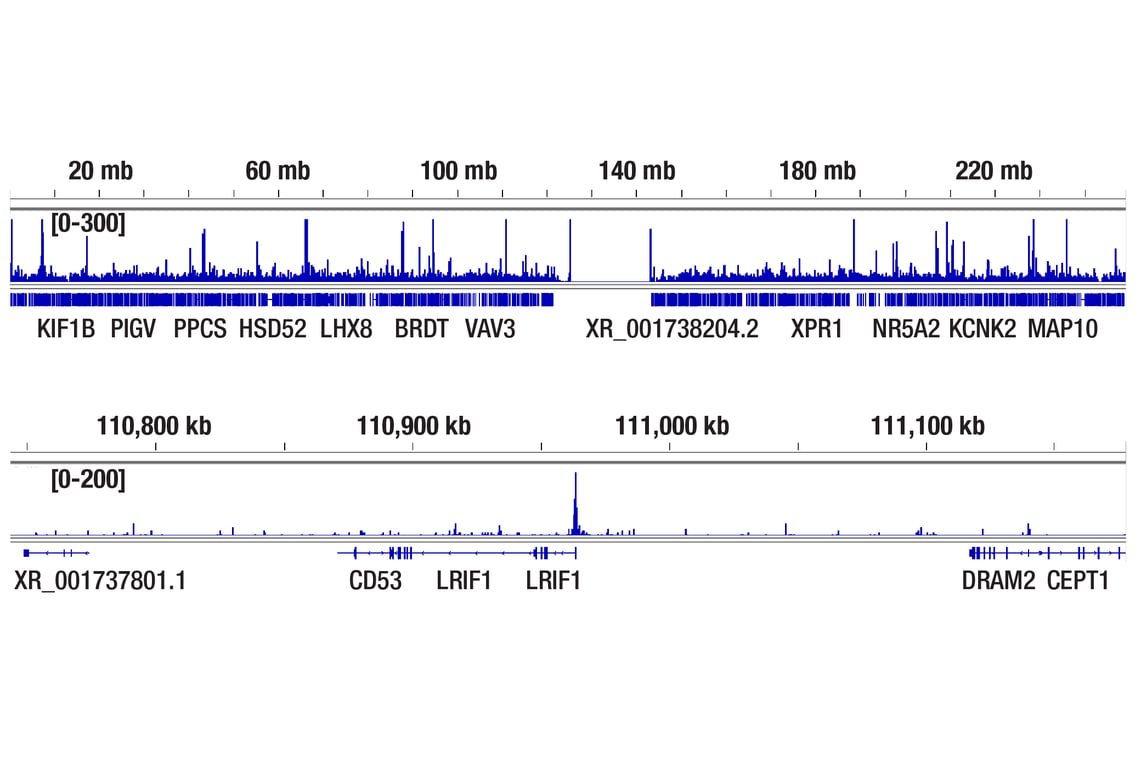


Monoclonal antibody is produced by immunizing animals with a synthetic peptide corresponding to the amino terminus of histone H4 in which Lys20 is tri-methylated.


Product Usage Information
For optimal ChIP and ChIP-seq results, use 10 μl of antibody and 10 μg of chromatin (approximately 4 x 106 cells) per IP. This antibody has been validated using SimpleChIP® Enzymatic Chromatin IP Kits.
The CUT&RUN dilution was determined using CUT&RUN Assay Kit #86652.
The CUT&Tag dilution was determined using CUT&Tag Assay Kit #77552.
| Application | Dilution |
|---|---|
| Western Blotting | 1:1000 |
| Chromatin IP | 1:50 |
| Chromatin IP-seq | 1:50 |
| CUT&RUN | 1:50 |
| CUT&Tag | 1:50 |



Specificity/Sensitivity
Species Reactivity:
Human, Mouse, Rat, Monkey




Supplied in 10 mM sodium HEPES (pH 7.5), 150 mM NaCl, 100 µg/ml BSA, 50% glycerol and less than 0.02% sodium azide. Store at –20°C. Do not aliquot the antibody.


参考图片
CUT&Tag was performed with HeLa cells and Tri-Methyl-Histone H4 (Lys20) (D84D2) Rabbit mAb, using CUT&Tag Assay Kit #77552. DNA library was prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figures show binding across chromosome 1 (upper), including the LRIF1 gene (lower).
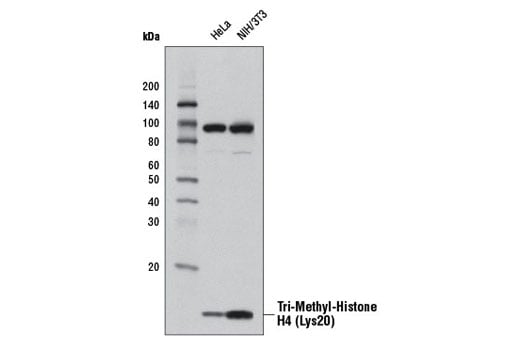
Western blot analysis of extracts from HeLa and NIH/3T3 cells using Tri-Methyl-Histone H4 (Lys20) (D84D2) Rabbit mAb.
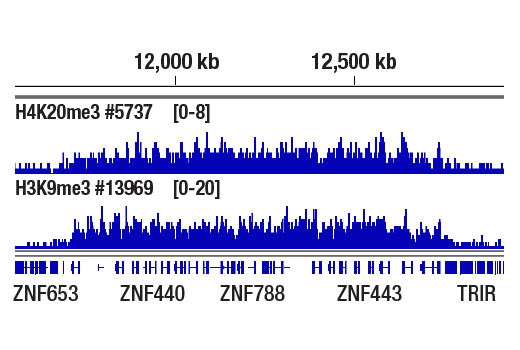
Chromatin immunoprecipitations were performed with cross-linked chromatin from HeLa cells and either Tri-Methyl-Histone H4 (Lys20) (D84D2) Rabbit mAb or Tri-Methyl-Histone H3 (Lys9) (D4W1U) Rabbit mAb #13969, using SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA Libraries were prepared using SimpleChIP® ChIP-seq DNA Library Prep Kit for Illumina® #56795. H4K20me3 and H3K9me3 are known to have similar binding pattern on chromatin. The figure shows binding of both H4K20me3 and H3K9me3 across ZNF genes, known target genes of both H4K20me3 and H3K9me3.
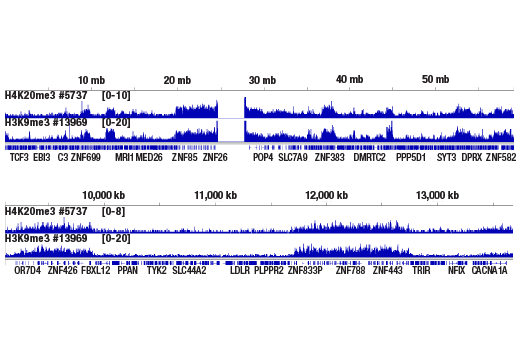
Chromatin immunoprecipitations were performed with cross-linked chromatin from HeLa cells and either Tri-Methyl-Histone H4 (Lys20) (D84D2) Rabbit mAb or Tri-Methyl-Histone H3 (Lys9) (D4W1U) Rabbit mAb #13969, using SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA Libraries were prepared using SimpleChIP® ChIP-seq DNA Library Prep Kit for Illumina® #56795. H4K20me3 and H3K9me3 are known to have similar binding pattern on chromatin. The figure shows binding of both H4K20me3 and H3K9me3 across chromosome 19 (upper), including ZNF genes (lower), known target genes of both H4K20me3 and H3K9me3.
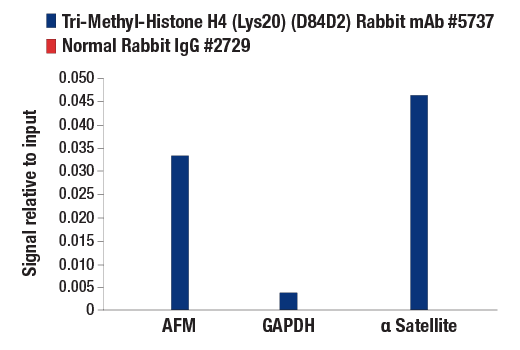
Chromatin immunoprecipitations were performed with cross-linked chromatin from HeLa cells and either Tri-Methyl-Histone H4 (Lys20) (D84D2) Rabbit mAb or Normal Rabbit IgG #2729, using SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. The enriched DNA was quantified by real-time PCR using SimpleChIP® Human α Satellite Repeat Primers #4486, SimpleChIP® Human AFM Intron 1 Primers #5098, and SimpleChIP® Human GAPDH Exon 1 Primers #5516. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin, which is equivalent to one.
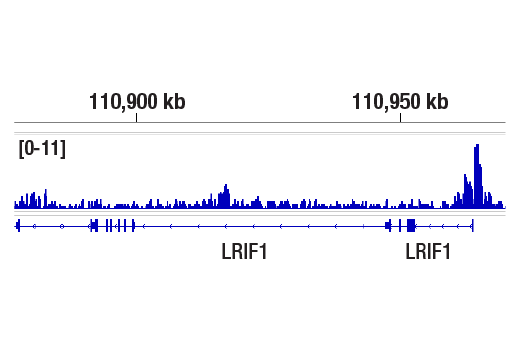
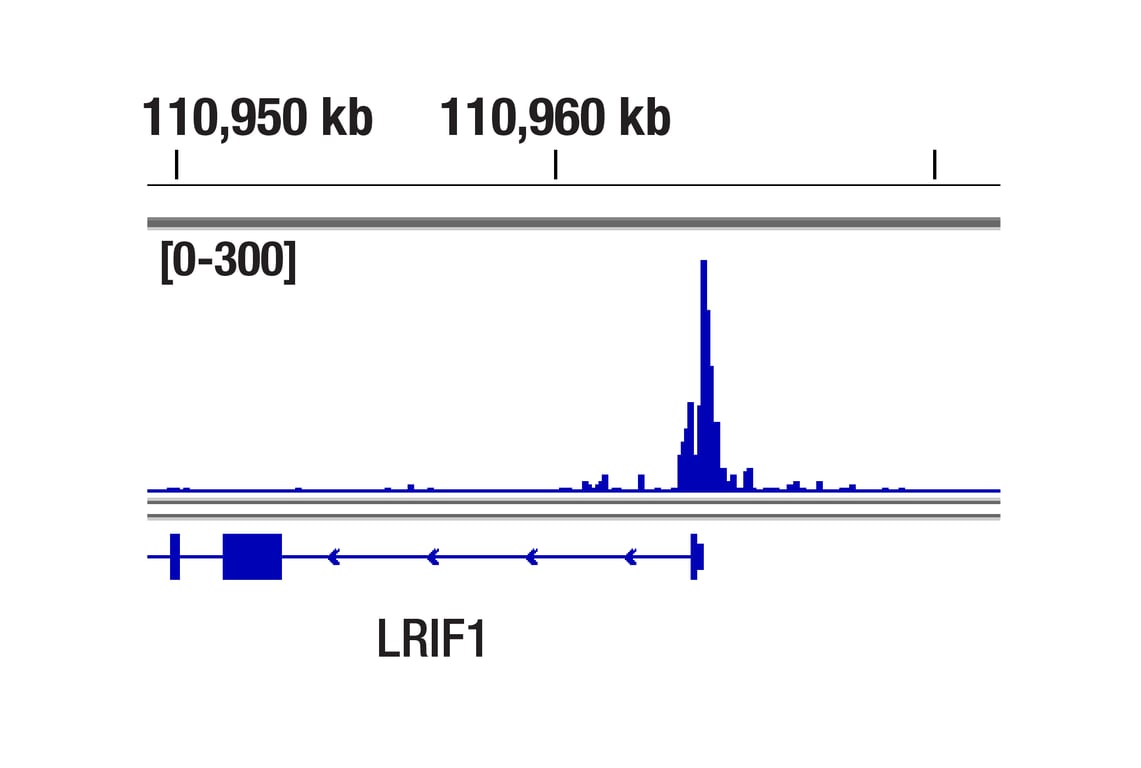
CUT&RUN was performed with HeLa cells and Tri-Methyl-Histone H4 (Lys20) (D84D2) Rabbit mAb, using CUT&RUN Assay Kit #86652. DNA library was prepared using DNA Library Prep Kit for Illumina Systems (ChIP-seq, CUT&RUN) #56795. The figures show binding across LRIF1, a known target gene of Tri-Methyl-Histone H4 (Lys20) (see additional figure containing CUT&RUN-qPCR data).
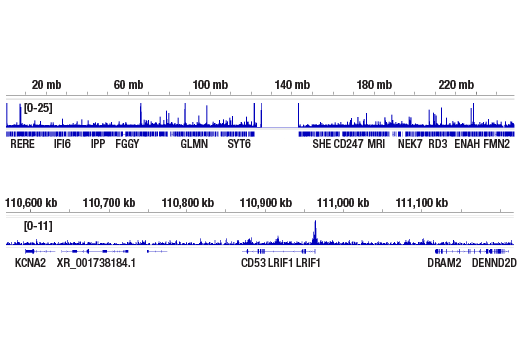
CUT&RUN was performed with HeLa cells and Tri-Methyl-Histone H4 (Lys20) (D84D2) Rabbit mAb, using CUT&RUN Assay Kit #86652. DNA library was prepared using DNA Library Prep Kit for Illumina Systems (ChIP-seq, CUT&RUN) #56795. The figures show binding across chromosome 1 (upper), including LRIF1 (lower), a known target gene of Tri-Methyl-Histone H4 (Lys20) (see additional figure containing CUT&RUN-qPCR data).
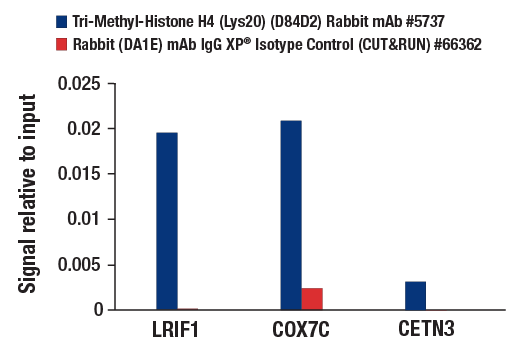
CUT&RUN was performed with HeLa cells and treatment and either Tri-Methyl-Histone H4 (Lys20) (D84D2) Rabbit mAb without product number or Rabbit (DA1E) mAb IgG XP® Isotype Control (CUT&RUN) #66362, using CUT&RUN Assay Kit #86652. The enriched DNA was quantified by real-time PCR using human LRIF1 promoter primers, human COX7C promoter primers, and human CETN3 promoter primers. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin, which is equivalent to one.
CUT&Tag was performed with HeLa cells and Tri-Methyl-Histone H4 (Lys20) (D84D2) Rabbit mAb, using CUT&Tag Assay Kit #77552. DNA library was prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figure shows binding across the LRIF1 gene.






 用小程序,查商品更便捷
用小程序,查商品更便捷